User story: Unraveling the behavior of the hIDO1 protein
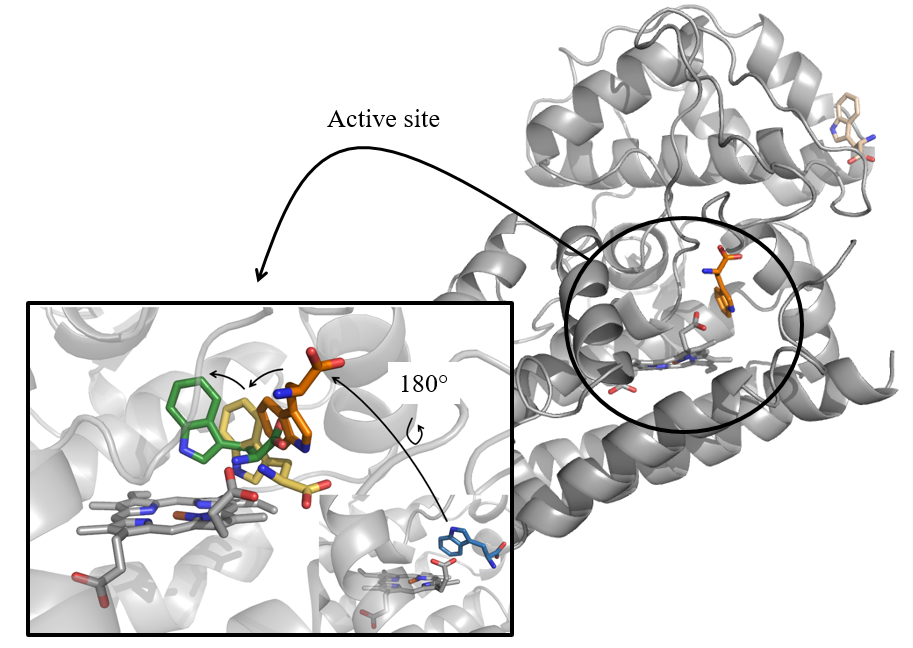
Thanks to computer simulations, the behaviour of the dynamic loop of hIDO1 during substrate positioning was unravelled.
Thanks to a multidisciplinary approach involving crystallographic studies and simulation performed on CECI’s supercomputers, Manon Mirgaux (University of Namur) was able to unravel the dynamic behaviour of the protein hIDO1. This protein is a target for immunotherapy, and understanding its dynamism is a key ingredient towards the design of new drugs, including the treatment of cancer.
The mechanisms of cancer
Cancer is one of the most common causes of death in the world, which involves an abnormal cell growth. Normally, after a given time fulfilling its goal, a cell is supposed to die in order to make room for new ones. This is a natural and controlled mechanism, which involves (chemical) communication between the different partners (other cells, immune system, etc.) to work in harmony. Thus, if a cell is malfunctioning due to a mistake in its DNA, either the mistake is fixed, or if it is beyond repair, the cell is asked to self-destruct. For different reasons, it may happen that a cell is not responding correctly to the signals or the fixing mechanism is broken, thus other partners have no control on this cell. It can start to reproduce without any control, thus resulting in abnormal cell growth. Further complications arise if the cells acquire the capacity to spread to other parts of the body (metastasis).
A new therapeutic target: hIDO1
Every cell function is actually possible thanks to proteins. More precisely, most of the chemical reactions inside the cell are mediated by a kind or protein, called an enzyme. Turning one biological molecule into another generally involves a chain of chemical reactions (a pathway), each catalysed (helped) by a different enzyme. For example, turning a molecule of sugar into energy that the cell can actually use involves about 20 of those reactions. But in cancer, it may be that one kind of enzyme is suractivated, which affects the pathway where it is involved, and thus, the overall behaviour of the cell. Therefore, inhibition of such proteins (therapeutic targets) is an important part of cancer research.
 The human Indoleamine 2,3-dioxygenase 1 (hIDO1) is a protein involved in the thryptophane-kynurenine pathway [1]. Its suractivation is linked to induce immune escape and resistance to immunotherapy. Manon Mirgaux, from the University of Namur studies this protein. For the design of new inhibitors, it is essential to understand the full structure of the protein including the interaction with its substrate (drugs). The usual technique is crystallography, which gives a snapshot of the position of the atoms inside the protein. But in hIDO1, its dynamic loop, which involved in the substrate positioning and therefore dictating its response to drugs is still unclear. The reason is that difficult to observe this part due to its dynamic (disordered) nature. In other words, if one sees crystallography as a technique which gives a picture of the protein, such dynamic parts would be blurry on the photograph!
The human Indoleamine 2,3-dioxygenase 1 (hIDO1) is a protein involved in the thryptophane-kynurenine pathway [1]. Its suractivation is linked to induce immune escape and resistance to immunotherapy. Manon Mirgaux, from the University of Namur studies this protein. For the design of new inhibitors, it is essential to understand the full structure of the protein including the interaction with its substrate (drugs). The usual technique is crystallography, which gives a snapshot of the position of the atoms inside the protein. But in hIDO1, its dynamic loop, which involved in the substrate positioning and therefore dictating its response to drugs is still unclear. The reason is that difficult to observe this part due to its dynamic (disordered) nature. In other words, if one sees crystallography as a technique which gives a picture of the protein, such dynamic parts would be blurry on the photograph!
Simulation of the dynamic loop of hIDO1
In order to capture the motion of this loop during substrate entrance, Molecular Dynamics (MD) simulations are essential. MD simulates the changes in positions of the atoms of a protein over time. In practice, starting from different crystallographic structures with different intermediates of the substrate, a full model has been reconstructed to complete the missing information on the loop. Then, simulations are launched to see the evolution of the loop during substrate positioning.
The benefit of this approach is unquestionable, since it complements the crystallographic observations, rather frozen (fixed), with an idea of the true protein's behaviour in vivo. These results allow making crucial advances in the understanding of this enzyme for the design of new drugs for, e.g., the treatment of cancer.
HPC to the rescue!
In molecular dynamics, the movement of each atom over a given period of time, sufficient to capture the phenomenon, is computed and then analysed. One simulation occurs in 1 month whereas it would have taken more than a year on a personal computer.
Manon: “The assistance of CECI's logistics team was crucial to the successful completion of the simulations and the provision of sound advice.”
Links
- Related publication: "Temporary Intermediates of L-Trp Along the Reaction Pathway of Human Indoleamine 2,3-Dioxygenase 1 and Identification of an Exo Site", by Manon Mirgaux, Laurence Leherte and Johan Wouters (University of Namur), published in the International Journal of Tryptophan Research in 2021 (DOI: 10.1177/11786469211052964).
- Twitter accounts: Manon Mirgaux, Johan Wouters' lab and University of Namur.
- [1]: The kynurenine pathway is a sequence of reaction which turns the tryptophane, an amino-acid (building blocks of proteins), into Nicotinamide Adenine Dinucleotide (NAD+), involved in important biological functions such as respiration.
About Manon Mirgaux
 Manon Mirgaux completed her master’s thesis on the human Phosphoserine Phosphatase (hPSP) in 2019. She is is currently pursuing a PhD thesis under the supervision of Prof. Johan Wouters in the CBS laboratory of the University of Namur. Her fields of interest go from crystallography to simulation of proteins, including the design of new inhibitors.
Manon Mirgaux completed her master’s thesis on the human Phosphoserine Phosphatase (hPSP) in 2019. She is is currently pursuing a PhD thesis under the supervision of Prof. Johan Wouters in the CBS laboratory of the University of Namur. Her fields of interest go from crystallography to simulation of proteins, including the design of new inhibitors.