User story: Understanding how radio waves propagate in the head with Shamo
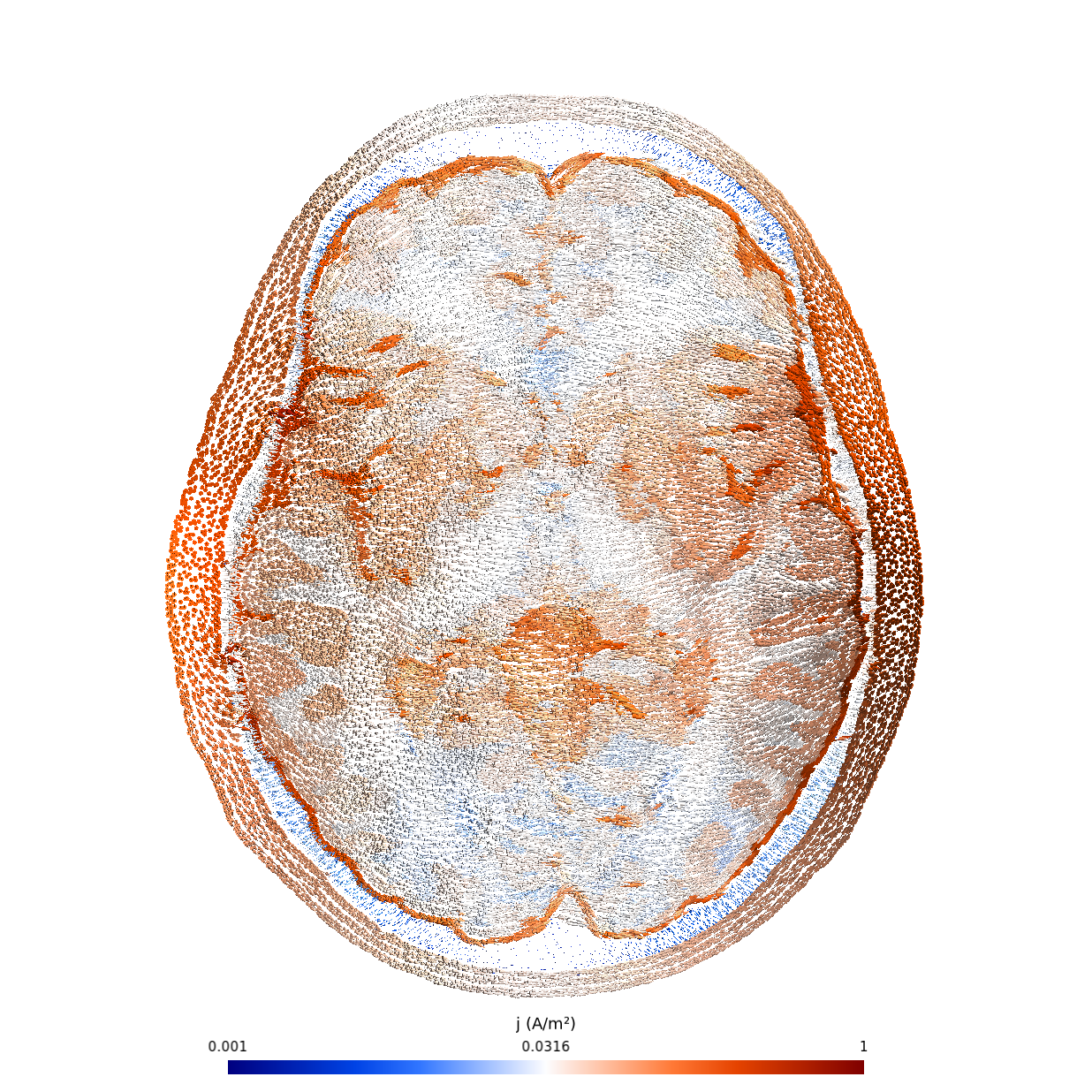
The effect of the application of a current by electrodes in a tDCS experiment, as computed thanks to the Shamo package.
Martin Grignard (University of Liège) developed new simulation software, called Shamo. This package smoothly interfaces with existing tools to easily simulate the propagation of electromagnetic waves through the brain. Indeed, while this propagation is the basis of different medical techniques, interpreting the data correctly requires to take into account the complexity and uniqueness of each head.
Understanding the brain
The brain is one of the most complex parts of the human or animal body. It contains billions of neurons, interconnected with each other, which give rise to our everyday movements and thoughts. Thus, understanding such complex machinery requires different techniques, such as functional magnetic resonance imaging (fMRI), electroencephalography (EEG) or others. While the first method measures the activity by targeting the blood flows in the brain, the latter actually records its electrical activity via electrodes placed on the head of the subject, which measure the variation of electric potential. EEG is typically non-invasive, and can be used to diagnose disorders such as epilepsy.
However, knowing from which part of the brain the signal is coming from is not a simple task. Indeed, from its source to its destination, the signal (which is nothing but an electromagnetic wave) passes through different media (brain tissues, bones, skin, etc.), and each of them propagates the wave differently because they have different physical properties (such as electrical conductivity or permittivity [1]). It is the same effect that one encounters with a straw in a glass of water: while the straw remains straight, the trajectory of the light is modified by the liquid, so that it appears bent. Thus, to know the exact source of the signal, practitioners should account for the different tissues and they have to do it for each subject since every head is different.
Shamo: one tool to rule them all
In practice, to determine the source of a signal, one needs to compute a so-called forward model (a mathematical function), which allows mapping a change of potential measured by the electrode to the location of its source. Such model is determined through the simulation of the propagation of the signal in a head, simulated by cutting a model of this head in small pieces with a given set of properties (the simulation time, but also the quality of the results, depends on how fine this division is) and propagating the wave from its source in each element. An example is given below:
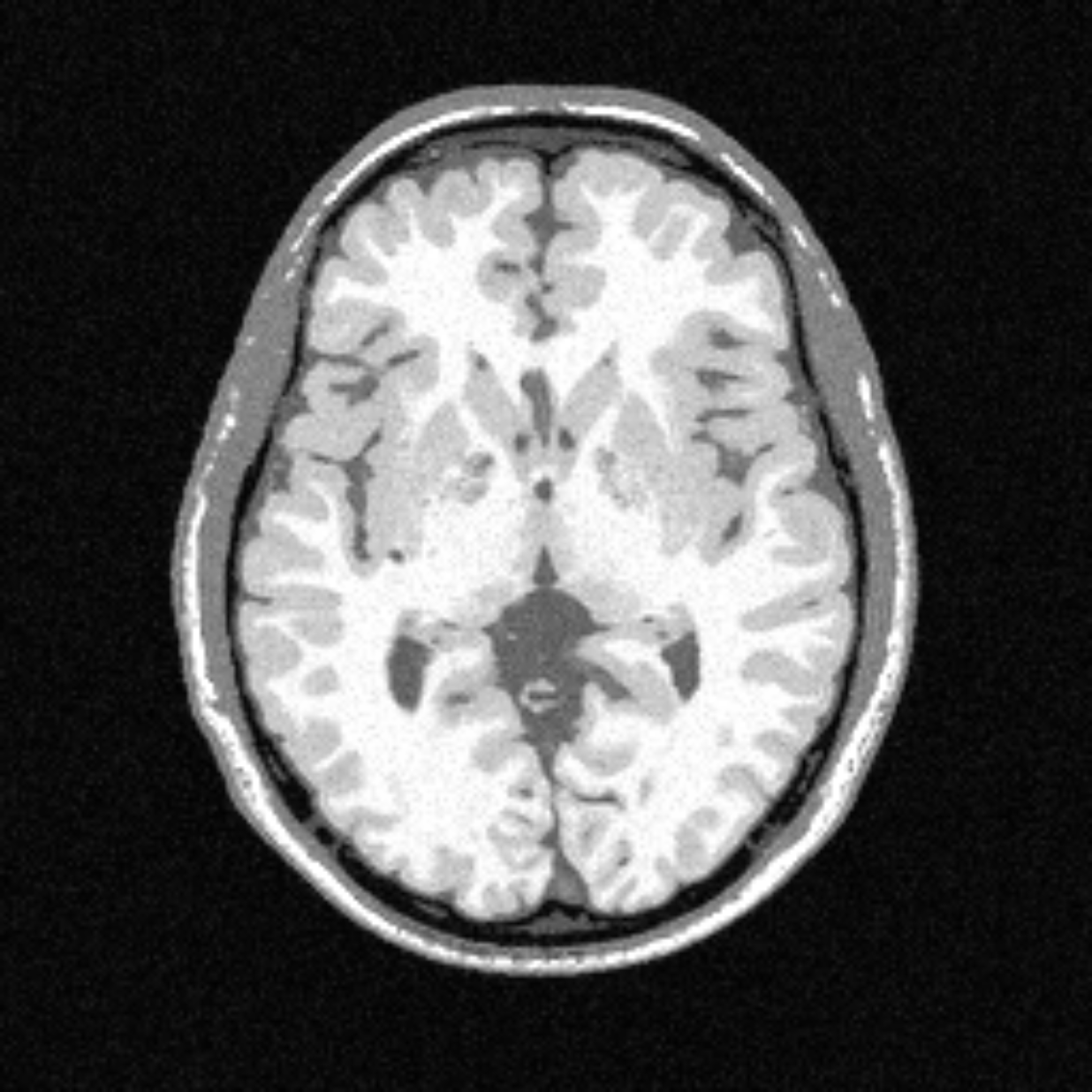
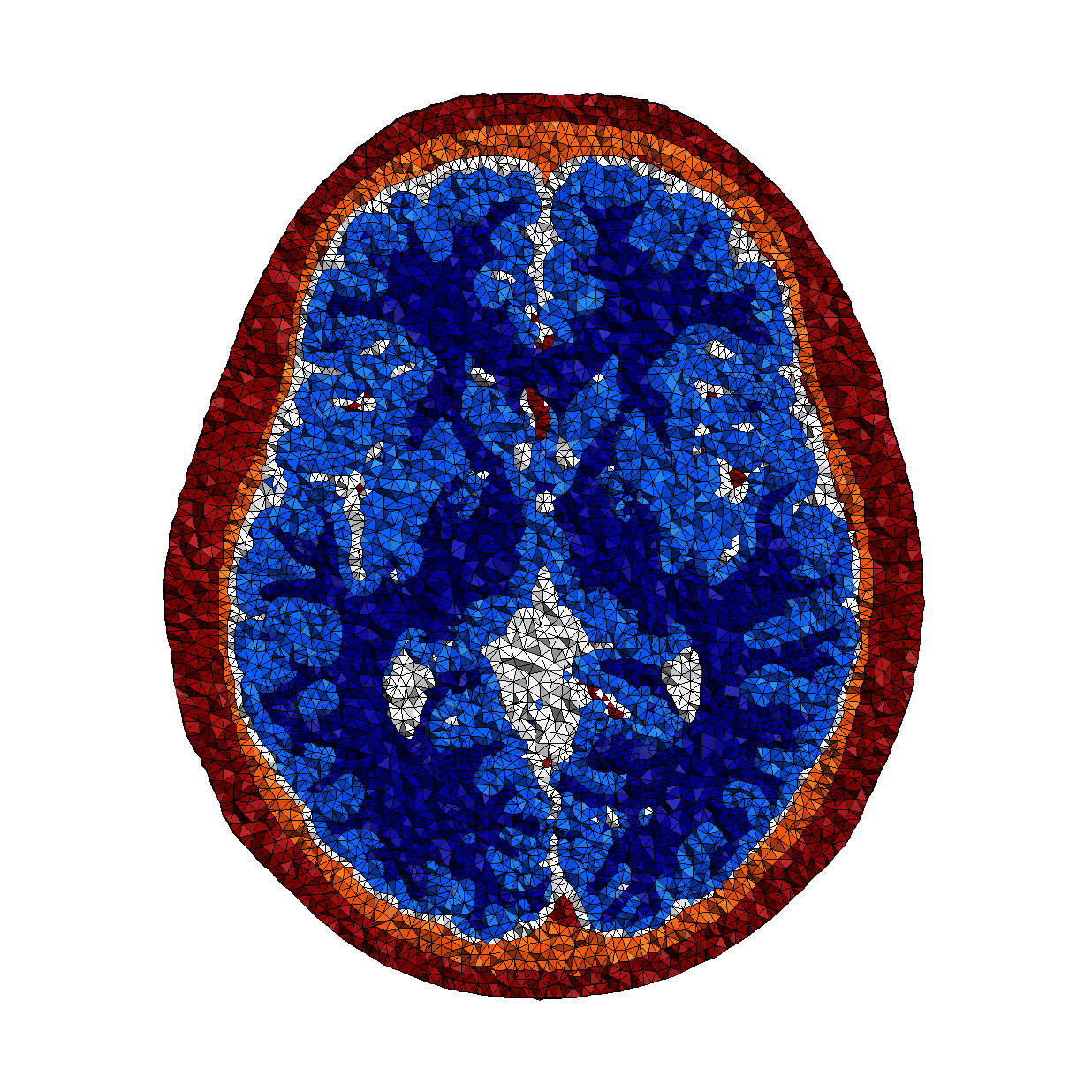
Head model (right) constructed from a MRI image (left) and discretized into small pieces, each with a given electrical conductivity (represented in the image by the different colors).
However, in order to build the forward model, a simulation must be performed for each electrode (there can be up to 256 of them!). Moreover, the said physical properties of the tissues are hard to determine accurately and have been shown to vary widely in between subjects. To account for this variability, one can build several forward models with different sets of properties by repeating the aforementioned process.
Furthermore, the different programs required to perform such simulations (from cutting the head model to the actual propagation of the wave inside this model and further post-treatments) are difficult to use together without proper knowledge.
Thankfully this problem is the subject of the PhD thesis of Martin Grignard (from the GIGA research center of the University of Liège), which developed an end-to-end solution called Shamo (to be pronounced [ʃɑ:mɔ:]) which is a friendly interface to the aforementioned programs, written in a powerful scripting language, Python.
Supercomputing to the rescue!
The goal of Shamo is to smooth the process, which can now be easily applied to multiple simulations. Thanks to the power of supercomputing , and since each simulation is independent, they can run at the same time [2]. This allows researchers to better account for the different sources of variability and thus leads to more cautious analysis and nuanced interpretations.
Furthermore, Martin is also interested to derive surrogate (i.e., simplified) models from its simulations. This is now easier given that a lot of parameter sets can be tested at the same time. The programs also generate a lot of data, which can now be handled and treated efficiently.
Martin Grignard: “To provide some figures [for our last article], we built 6 EEG surrogate models, each based on 100 EEG forward models which required 59 simulations, each taking around 15 min. Sequentially, this would have taken a bit more than a year. We ran 100 simulations at a time on one of the CECI clusters, thus reducing the time to less than a week.”
With Shamo, it is now easier to build either EEG models (locating the source of the signal), but also to simulate the reverse experience, called transcranial direct-current stimulation (tDCS), where the electrodes are used to stimulate a specific part of the brain, considering, again, the uncertainty on the physical properties of the tissues.
Links
- The source code of Shamo and its documentation.
- A recent publication: “Shamo: A Tool for Electromagnetic Modeling, Simulation and Sensitivity Analysis of the Head” (also available on their institutional repository), by Martin Grignard, Christophe Geuzaine and Christophe Phillips, published in Neuroinfomatics in 2022.
- Twitter accounts: the GIGA research center, and the University of Liège.
- [1]: The permittivity is a quantity indirectly related to the propagation of a wave into a medium: the larger the permittivity, the less the wave is propagated. The anisotropy, the fact that the wave may propagate easier in a given direction than in another, is also important in this kind of simulation.
- [2]: This is the so-called embarrassingly parallel paradigm, where little or no effort is required to separate the problem into a number of parallel tasks, thanks to a small number of dependencies ... Actually the best case scenario for any complex calculation!
About Martin Grignard
 In 2012, he first started to learn art for a year. But he quickly came back to his first love: Science, and thus studied industrial engineering for 5 years at the Gramme Institute (Liège, Belgium). Currently, he is a PhD student at the CRC GIGA In-vivo Imaging lab (Liège, Belgium).
In 2012, he first started to learn art for a year. But he quickly came back to his first love: Science, and thus studied industrial engineering for 5 years at the Gramme Institute (Liège, Belgium). Currently, he is a PhD student at the CRC GIGA In-vivo Imaging lab (Liège, Belgium).